In this article, I briefly describe restriction enzymes, their recognition sequences, and the frequency of recognition sequences.
What are restriction enzymes?
The enzyme that cleaves DNA at specific recognition sites, is called a restriction enzyme. These belong to the endonuclease group of enzymes. Identifying and manipulating restriction endonucleases in the 1960s and early 1970s made DNA cloning a reality. Restriction enzymes make two incisions, once through each sugar-phosphate backbone of the DNA double helix.
Restriction enzymes protect bacteria against invading viruses. These enzymes selectively cut foreign DNA in a process called restriction digestion. Methyltransferase, a modification enzyme modifies the prokaryotic DNA, thus blocking cleavage. In this way, host DNA is protected by methyltransferase, and these two processes form the restriction-modification system. This modification renders the host DNA resistant to degradation by the endonuclease. Three different classes of restriction endonucleases are recognized, i.e., type I, type II, and type III. Types I and III are more complex than type II and have only a very limited role in genetic engineering. Type II restriction endonucleases are the cutting enzymes that are important in gene cloning.
One restriction endonuclease- one recognition sequence
There is a specific recognition sequence for each restriction endonuclease at which it cuts a DNA molecule. Each endonuclease will cleave DNA only at the recognition sequence. Different enzymes vary in recognizing target sites. Many endonucleases recognize hexanucleotide target sites, whereas some recognize four, five, or even eight nucleotide sequences. Sau3a from Staphylococcus aureus strain 3A recognizes GATC, and AluI from Arthrobacter luteus cuts at AGCT. Some enzymes with degenerate recognition sequences cut DNA at any one of a family of related sites. Hinf1 from Haemophilus influenzae strain Rf, for instance, recognizes GANTC, so cuts at GAATC, GATTC, GAGTC, and GACTC.
Blunt ends and sticky ends
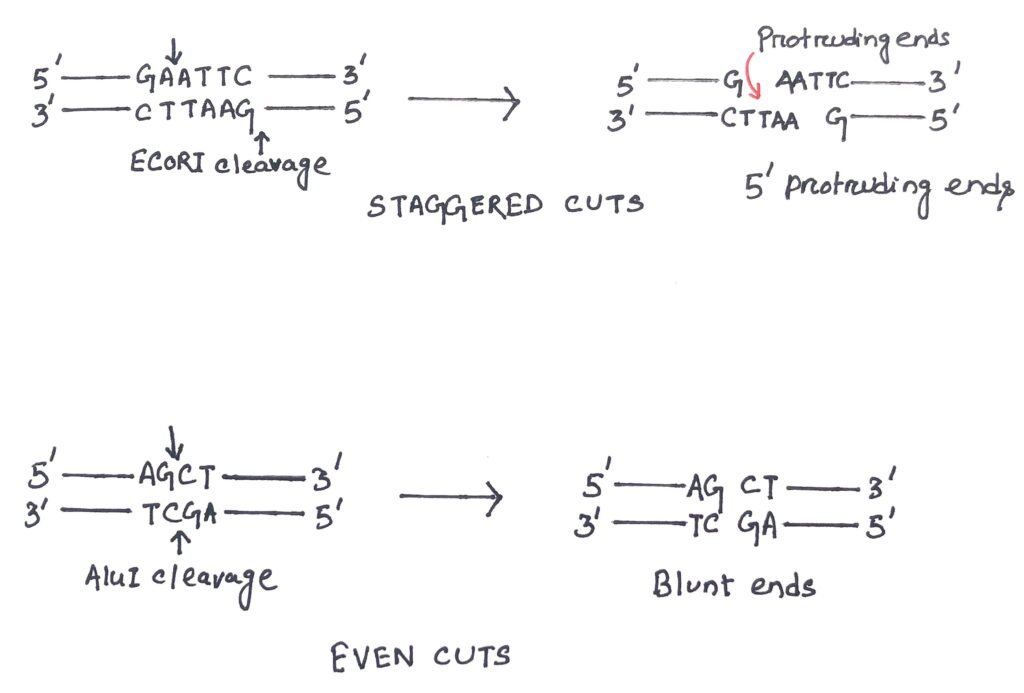
In a gene cloning experiment, the cut produced by a restriction endonuclease is of considerable importance. A blunt-ended DNA is the simplest DNA end of a double-stranded molecule. In a blunt-ended molecule, both strands terminate in a base pair (figure 1). Many restriction endonucleases make a simple double-stranded cut in the middle of the recognition sequence, resulting in a blunt end. The blunt end does not always have the advantages as during the sub-cloning experiment, it has the disadvantage of potentially inserting the insert DNA in the opposite orientation desired. Blunt ends are always compatible with each other. The restriction enzymes PuvII and AluII are blunt end cutters.
A stretch of unpaired nucleotides at the end of a DNA molecule makes an overhang. Various overhangs create non-blunt ends. These unpaired nucleotides can be in any one of the two strands, creating either 3′ or 5′ protruding ends (figure 1). In most of the cases, these overhangs are palindromic.
Sticky ends
Longer overhangs are called cohesive ends or sticky ends. Many restriction endonucleases cut DNA in a different way. Mostly, they cut the two DNA strands four base pairs from each other, creating a four-base 5′ overhang in one molecule and a complementary 5′ overhang in another. These ends are easily joined back together, thus called as cohesive ends.
Different restriction endonucleases usually create different overhangs. Thus, it is possible to cut a piece of DNA with two different enzymes and then join it with another DNA molecule with ends created by the same enzymes. Since the overhangs have to be complementary for the ligase to work, the two molecules can only join in one orientation. Some restriction endonucleases with different recognition sequences may produce the same sticky ends, e.g., BamHI (recognition sequence GGATCC) and BglII( recognition sequence AGATCT) both produce GATC sticky ends. The endonuclease Sau3A also produces the same sticky end, which recognizes only the tetranucleotide GATC (figure 2).

Frequency of recognition sequences
In a DNA molecule, the number of recognition sequences for a specific restriction endonuclease can be calculated mathematically. A tetranucleotide sequence (e.g., GATC) should occur once every 44 = 256 nucleotides, and a hexanucleotide sequence (e.g., GGATCC) once every 46=4096 nucleotides. Restriction sites are not evenly spaced along a DNA molecule. The fragments produced by cutting λ DNA with BglII, BamHI, and SalI are not equal in size.
Restriction digests
Restriction enzymes are used to digest plasmid or genomic DNA using commercial enzymes and buffer solutions. All restriction enzymes require Mg2+, at a concentration of up to 10 mM, but they require different pHs, NaCl concentrations, or other solution constituents for optimum activity. The digestion of a sample plasmid with two different restriction enzymes, BamHI and EcoRI shows fragments of different sizes (figure 3). A restriction digest will result in several DNA fragments, the sizes of which depend on the exact positions of the recognition sequences for the endonuclease in the original molecule. The process of gel electrophoresis helps to determine the number and sizes of the DNA fragments after restriction digestion.

Conclusion
Restriction enzymes, belonging to the endonuclease group, cleave DNA at specific recognition sites. These enzymes selectively cut foreign DNA in a process called restriction digestion. Three different classes of restriction endonucleases are recognized, i.e., type I, type II, and type III. Each endonuclease will cleave DNA only at the recognition sequence. Different enzymes vary in recognizing target sites.
Many restriction endonucleases make a simple double-stranded cut in the middle of the recognition sequence, resulting in a blunt end. Blunt ends are always compatible with each other. The restriction enzymes PuvII and AluII are blunt end cutters. Many restriction endonucleases cut DNA differently. Mostly, they cut the two DNA strands four base pairs from each other, creating a four-base 5′ overhang in one molecule and a complementary 5′ overhang in another. These ends are easily joined back together, thus called cohesive ends or sticky ends.
A restriction digest will result in several DNA fragments, the sizes of which depend on the exact positions of the recognition sequences for the endonuclease in the original molecule.
You may also like:

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around twelve years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.
