In this article, I briefly describe the preparation of plasmid DNA.
Plasmid DNA
A plasmid is a small, circular, extrachromosomal DNA molecule in bacteria, which replicates independently. Chromosomes contain all the essential genetic information, whereas plasmids carry some additional genes, which aid in the survival of the organism. Plasmids provide some selective advantages such as antibiotic resistance to the organism. A plasmid possesses an origin of replication, which helps in independent replication. The preparation of plasmid DNA involves the same procedures as total cell DNA. However, in bacterial plasmid DNA preparation, it is essential to separate the plasmid DNA from the chromosomal DNA.
Plasmids and chromosomal DNA differ in conformation. Plasmids are circular DNA molecules, which break into linear fragments during preparation of the cell extract. A method of separating circular from linear molecules will therefore result in pure plasmids. The separation can be carried out by size-based separation and conformation-based separation.
Size-based separation
The process of bacterial cell disruption is carried out very gently. To prevent the cell from bursting, it is treated with EDTA and lysozyme in the presence of sucrose. Partially wall less cells, also called sphaeroblasts are formed that retain an intact cytoplasmic membrane (figure 1). The addition of a non-ionic detergent, Triton X-100, induces cell lysis with minimal breakage of the bacterial DNA. Then, it undergoes centrifugation, which leaves a cleared lysate, consisting almost entirely of plasmid DNA. However, a clear lysate will invariably retain some chromosomal DNA. Thus, size fractionation does not aid in the entire removal of contaminants. So, alternative methods are taken into consideration.

Conformation based separation
The supercoiled plasmids are easily separated from the non-supercoiled DNA by conformation-based separation. The two different types of conformation-based separation are alkaline denaturation and EtBr-CsCl density gradient centrifugation.
Alkaline denaturation
Non-supercoiled DNA is denatured at a narrow pH range. An increased pH disrupts the hydrogen bonding in non-supercoiled DNA molecules. the pH of a cell extract or a cleared lysate is increased up to 12.0-12.5 by the addition of NaOH, which disrupts the hydrogen bonding in non-supercoiled DNA molecules. The disruption of hydrogen bonding causes the unwinding of the DNA double helix and finally two polypeptide chains are separated (figure 2). With the addition of acid, these denatured DNA strands re-aggregate into a tangled mass. The insoluble network can be pelleted after centrifugation, leaving pure plasmid DNA in the supernatant. Under some circumstances ( cell lysis by SDS and neutralization with sodium acetate), most of the proteins and RNA also become insoluble and can be removed by centrifugation.

Ethidium bromide-cesium chloride(EtBr-CsCl) density gradient centrifugation
EtBr-Cscl density gradient centrifugation is a very efficient method for obtaining pure plasmid DNA. This method can be used to separate supercoiled DNA from non-supercoiled molecules (Figure 3b). Ethidium bromide binds to DNA molecules by intercalating between adjacent base pairs, causing partial unwinding of the double helix. In a solution of cesium chloride (CsCl), when undergoes centrifugation, the CsCl molecules dissociate under high centrifugal force, and the heavy Cs+ atoms are forced towards the outer end of the tube. Thus, it forms a shallow density gradient (figure 3a). DNA molecules placed in this gradient will migrate to the point where they have the same density as the gradient. The point is called the isopycnic point.
Under centrifugation, macromolecules present in the CsCl solution form bands at distinct points in the gradient. The gradient is sufficient to separate types of DNA with slight differences in density due to differing (G+C) content, or physical form (e.g., linear versus circular molecules).
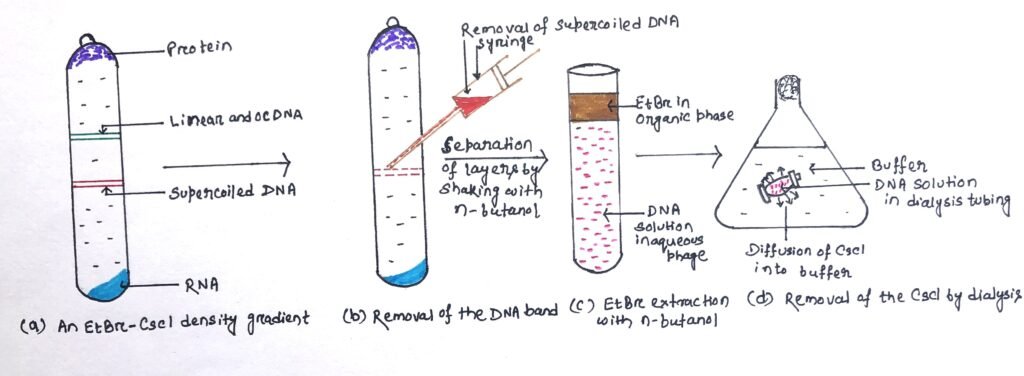
When a cleared lysate is subjected to this procedure, plasmids band at a distinct point, separated from the linear bacterial DNA, with the protein floating at the top of the gradient and RNA pelleted at the bottom. Under ultraviolet radiation, the position of the DNA bands can be visualized with fluorescein ethidium bromide. The EtBr bound to the plasmid DNA is extracted with n-butanol (figure 3c) and the CsCl is removed by dialysis (figure 3d). The resulting plasmid preparation is pure and can be used in cloning. Density gradient centrifugation can separate DNA, RNA, and protein and is an alternative to phenol extraction and ribonuclease treatment for DNA purification.
Conclusion
A plasmid is a small, circular, extrachromosomal DNA molecule in bacteria, which replicates independently. Plasmids are circular DNA molecules, which break into linear fragments during preparation of the cell extract. A method of separating circular from linear molecules will therefore result in pure plasmids. The separation is of two types, i.e. size-based separation and conformation-based separation.
The supercoiled plasmids are easily separated from the non-supercoiled DNA by conformation-based separation. The two different types of conformation-based separation are alkaline denaturation and EtBr-CsCl density gradient centrifugation. EtBr-Cscl density gradient centrifugation is a very efficient method for obtaining pure plasmid DNA. Density gradient centrifugation can separate DNA, RNA, and protein and is an alternative to phenol extraction and ribonuclease treatment for DNA purification.
You may also like:

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around twelve years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.
