In this article, I briefly describe the duplication of retrovirus in a host cell.
The enveloped virus-Retrovirus
These are enveloped viruses, that belong to the family Retroviridae. A retrovirus is an RNA virus duplicated in a host cell using the reverse transcriptase enzyme to produce DNA from its RNA genome. When the virus infects a host, the single-stranded RNA viral genome and the enzyme get an entry into the host cell. The enzyme reverse transcriptase catalyzes the synthesis of a DNA strand complementary to the viral RNA. Then, it degrades the RNA strand of the viral RNA-DNA hybrid and replaces it with DNA. An enzyme integrase helps in the incorporation of the resulting duplex DNA into the host’s genome. The virus thereafter replicates as part of the host cell’s DNA. The integrated viral genes can be activated and transcribed. The viral proteins with the viral genome itself packaged as new viruses.
Retrovirus- Structure
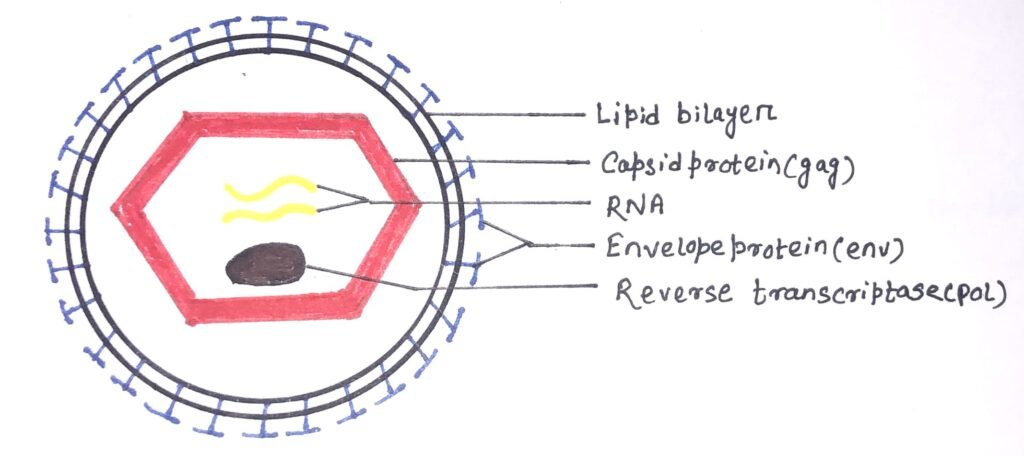
It consists of two concentric outer circles of the lipid bilayer, which has the envelope protein complex within it. Virions of retroviruses consist of enveloped particles about 100 nm in diameter. The components of a virion are the outer envelope, RNA, and protein.
Outer envelope
The outer envelope comprises lipids and glycoproteins (figure 1) encoded by the env gene obtained from the host plasma membrane during the budding process. The lipid bilayer works as a protection layer for the retrovirus.
RNA
Each virion consists of a dimer RNA (figure 1), which is polyadenylated at the 3′ end and capped at the 5′ end. There are terminal non-coding regions and internal regions present in the RNA genome. The terminal non-coding areas are important in the process of replication in the RNA genome and the internal regions encode virion proteins for gene expression. The copies of the RNA genome are in the form of a loop bound by nucleoproteins.
Protein
Retroviruses consist of gag proteins, protease, pol proteins, and env proteins (figure 1). The major component of the viral capsid is the Gag proteins, which are about 2000-4000 copies per virion. The transcript that contains gag and pol is translated into a long polyprotein. It is a single large polypeptide that is cleaved into six proteins with distinct functions. The expression of protease varies in different viruses. The pol gene encodes the protease that cleaves the long polypeptide into an integrase, and a reverse transcriptase. An integrase inserts the viral DNA into the host chromosome. The env gene encodes the proteins of the viral envelope.
There are long terminal repeat sequences (LTR) of a few hundred nucleotides in the viral linear RNA genome. These sequences are transcribed into the duplex DNA and facilitate the integration of the viral chromosome into the host DNA. The promoters for viral gene expression are present in these sequences. The retrovirus can bind to its target host cell by using the surface component of the env proteins. The membrane-anchored transmembrane component (TM) of the env facilitates the binding of the retrovirus to its target host cell.
Reverse transcriptase
It is also called RNA dependent DNA polymerase, a DNA polymerase enzyme, that transcribes single-stranded RNA into single-stranded DNA. The DNA dependent DNA polymerase activity of it synthesizes a second strand of DNA complementary to the reverse-transcribed single-stranded cDNA after degrading the original mRNA with its RNaseH activity. Reverse transcriptase was discovered by scientists Howard Temin and David Baltimore in the year of 1970.
HIV-1 reverse transcriptase from human immunodeficiency virus type 1, AMV reverse transcriptase from the avian myeloblastosis virus, M-MLV reverse transcriptase from the Moloney murine leukemia virus, etc are some well-known examples of reverse transcriptases.
The working of reverse transcriptase
Reverse-transcribing RNA viruses such as retroviruses use the enzyme during the replication process. They use it to reverse-transcribe their RNA genome into DNA, which is then integrated into the host genome and replicated along with it. The three different reactions catalyzed by reverse transcriptases are 1) RNA-dependent DNA synthesis, 2) RNA degradation, and 3) DNA-dependent DNA synthesis. Reverse transcriptase initiates DNA synthesis by a primer, a cellular tRNA obtained during an earlier infection and carried within the viral particle. This tRNA is base-paired at its 3′ end with a complementary sequence in the viral RNA. The new DNA strand is synthesized in the 5’→3′ direction. Reverse transcriptases, like RNA polymerases, do not have 3’→5′ proofreading exonucleases.
Reverse transcriptases have become important reagents in DNA cloning techniques. The synthesis of complementary DNA (cDNA) can be used to clone cellular genes. Reverse transcriptases have also been shown to be involved in processes such as transcript fusions, exon shuffling and creating artificial antisense transcripts. It is commonly used in research to apply the polymerase chain reaction technique to RNA in a technique called reverse transcription polymerase chain reaction (RT-PCR).
Reverse transcription of retroviruses
The genomes of retroviruses consist of two molecules of positive-sense single-stranded RNA with a 5′ cap and a 3′ polyadenylated tail. A retrovirus generates a provirus by reverse-transcription of the retroviral genome. An integrated provirus consists of a double-stranded DNA sequence. The integrated form (proviral) of all retroviruses contains transcription regulatory sequences primarily in Long Terminal Repeats (LTR). The sequences unique to the 5′ end and 3′ end of viral RNA (U5), and the repeated sequences at both ends of the viral RNA (R) give rise to the LTR sequences.
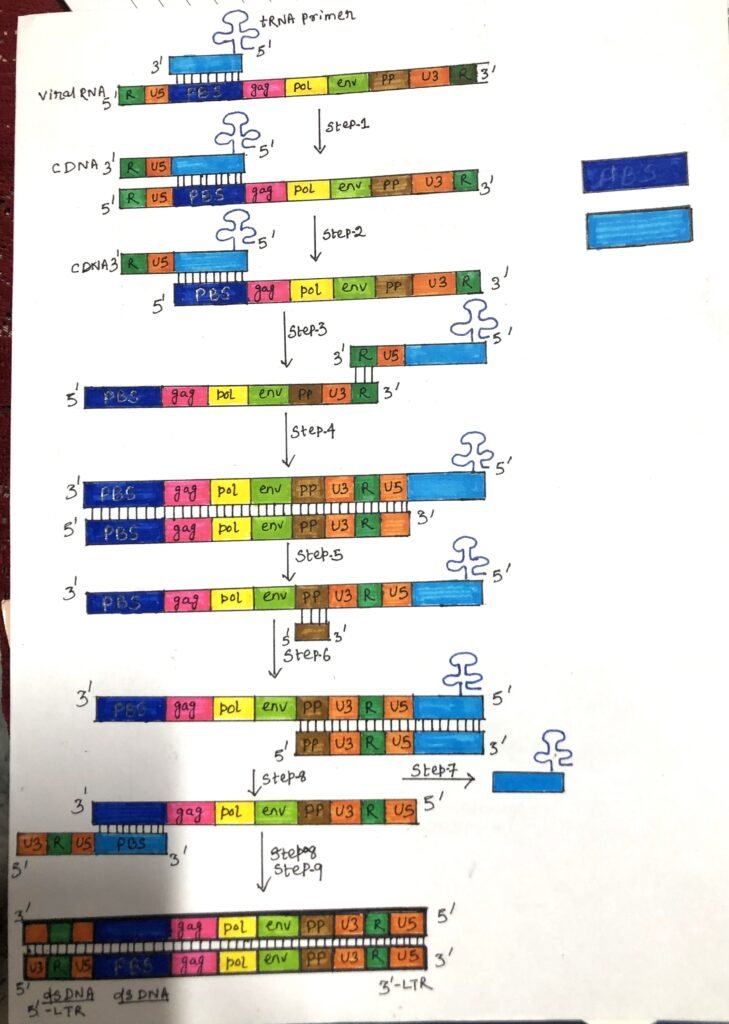
The procedure
- First, a tRNA primer binds to a site of 100 to 200 bases from the 5′ end at the primer binding site. Then, reverse transcriptase starts synthesis. (Step 1 of figure 2).
- Reverse transcriptase begins the synthesis of the first strand of cDNA (minus strand of DNA), which binds to the U5 (non-coding region) and R region (a direct repeat found at both ends of the RNA molecule) of the viral RNA. (Step 1 of figure 2).
- After copying of the U5 and repeat (R) regions, the nascent minus-strand DNA transfers the R sequences at the 3′ end of the same molecule of viral RNA, next to the poly(A) sequence. When the enzyme reaches the end, the 5′ terminal bases of RNA are degraded by RNAse H, which removes the U5 and R region, and as a result, the 3′ end of the DNA product is exposed (Step 2 of figure 2).
Jumping of the primer
- Then, the primer jumps to the 3′ end of the viral genome and the newly synthesized DNA strand hybridizes to the complementary R region on the RNA (Step 3 of figure 2).
- The first strand (the minus-strand DNA) of complementary DNA (cDNA) is extended. After the minus-strand DNA is elongated through the R U3 regions (Step-4 of figure-2), a reverse transcriptase RNase H cleaves the RNA template near its 3′ end just after a polypurine tract (ppt).
- The synthesis of the minus strand DNA or the first strand is completed. Then, synthesis of the second DNA strand or plus DNA strand is initiated from the viral RNA. The 3′ end of the viral RNA forms a primer for plus-strand DNA synthesis and it becomes a point for elongation (step-5 and step-6 of figure 2). The tRNA primer is removed and the majority of viral RNA is degraded by RNAse H leaving fragments to prime DNA synthesis (step-7 of figure 2).
- In the second jump, the primer binding site (PBS) from the second strand hybridizes with the complementary PBS on the first strand (step 8 of figure 2).
- Both strands are further extended (step 9 of figure 2) and can be incorporated into the host genome by the integrase enzyme.
Integration
The organization of proviral DNA and a transposon is same. At the target site, the provirus is flanked by short direct repeats of a sequence. Retroviral integrase enzyme inserts linear DNA directly into the host chromosome. Two base pairs of DNA are lost from each end of the retroviral sequence during the integration reaction. The enzyme integrase catalyzes all the stages of the integration reaction. In the viral DNA, integrase generates two base recessed at the 3′ ends in LTRs.
Integrase generates staggered 5′ ends in the target DNA. Finally, the enzyme attaches the 3′ ends of LTR with the staggered 5′ ends of target DNA to complete the integration reaction.
Strand transfer causing recombination
Double-stranded DNA production involves strand transfer, which causes recombination. Two copies of the RNA genomes, sometimes non-identical, are co-packaged in the genomes of retroviruses. The two strands of RNA undergo recombination, which provide a strong positive selective advantage for retroviruses. It allows them to repair breaks in the RNA and to exchange nucleic acid sequences.
The properties of the reverse transcriptase allow a transfer of the growing DNA strand between these genomes to occur occasionally at any point during reverse transcription. Thus, produce recombinant viral progeny. The virus promotes its structural diversity through recombination that helps it to resist host immunity and drug therapy. During the minus DNA strand synthesis in retrovirus, homologous recombination takes place.
Homologus recombination results from the growing point of reverse transcriptase, transferring to an identical sequence on the other RNA molecule of the dimer RNA. Homologous recombination can be the result of usual reverse transcriptase growing point transfer, called copy-choice. Or, it may be the result of an RNA break that forces the reverse transcriptase growing point to transfer, called forced copy-choice. In a complex multi-step invasion transfer mechanism, the second RNA template interacts with the growing DNA strand well behind the DNA 3′-terminus. The newly formed RNA-DNA hybrid expands by branch migration and eventually catches the elongating DNA primer 3′-terminus to complete the transfer.
The causal organism of cancer and AIDS- Retrovirus
Rretroviruses normally do not kill their host cells, rather remain integrated in the cellular DNA. They replicate, when the cell divides. Retroviruses, such as Rous sarcoma virus and mouse mammary tumor virus cause tumor growth. Rous sarcoma virus contains the src gene, a proto-oncogene that triggers tumor formation. Cancer can be triggered by proto-oncogenes that were mistakenly incorporated into proviral DNA or by the disruption of cellular proto-oncogenes. Rous sarcoma virus contains the src gene that triggers tumor formation. Non-transforming viruses can disrupt the expression of cell-cycle regulating proteins by inserting their DNa into proto-oncogene. The promoter of the provirus DNA can also cause over expression of regulatory genes.
The human immunodeficiency virus (HIV), which causes AIDS (Acquired immunodeficiency syndrome), is a retrovirus. It has an RNA genome with standard retroviral genes along with several other unusual genes (figure 3).
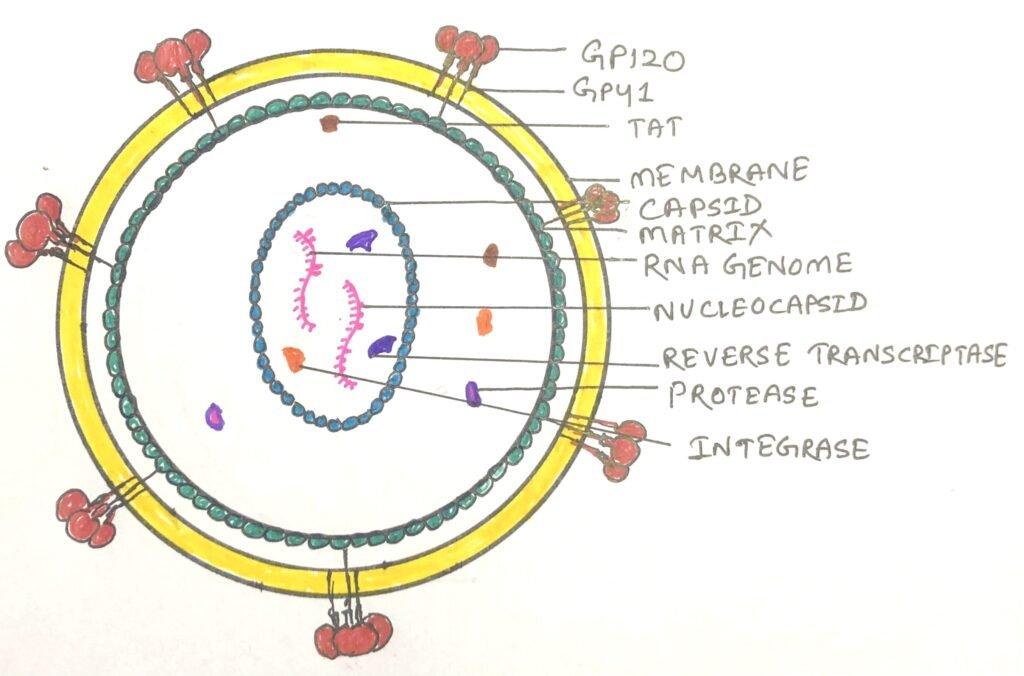
The HIV kills many of the cells it infects rather than causing tumor formation, which leads to suppression of the immune system in the host organism. Infection of HIV leads to the weak functioning of host immune system.
Conclusion
A retrovirus is an RNA virus duplicated in a host cell. It uses the reverse transcriptase enzyme to produce DNA from its RNA genome. The enzyme reverse transcriptase catalyzes the synthesis of a DNA strand complementary to the viral RNA. Then, it degrades the RNA strand of the viral RNA-DNA hybrid and replaces it with DNA. An enzyme integrase helps in the incorporation of the resulting duplex DNA into the host’s genome.
Retrovirus consists of two concentric outer circles of the lipid bilayer, which has the envelope protein complex within it. Reverse transcriptase was discovered by scientists Howard Temin and David Baltimore in the year of 1970. The DNA dependent DNA polymerase activity of it synthesizes a second strand of DNA complementary to the reverse-transcribed single-stranded cDNA after degrading the original mRNA with its RNaseH activity. A retrovirus generates a provirus by reverse-transcription of the retroviral genome.
The two strands of RNA undergo recombination, which provide a strong positive selective advantage for retroviruses. It allows them to repair breaks in the RNA and to exchange nucleic acid sequences. Retroviruses, such as Rous sarcoma virus and mouse mammary tumor virus cause tumor growth. Rous sarcoma virus contains the src gene, a proto-oncogene that triggers tumor formation. The human immunodeficiency virus (HIV) is a retrovirus, which causes AIDS (Acquired immunodeficiency syndrome).
You may also like

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.



