In this article, I briefly describe how the analysis of protein-protein interactions helps to understand protein function.
Understanding Protein Roles through Molecular Associations
The function of a specific protein can often be inferred by identifying the cellular components it binds to. When it comes to protein–protein interactions, associating a protein of unknown function with a well-characterized one can strongly suggest a functional link. A wide range of techniques is employed to study these interactions.
Protein Complexes are Purified
Researchers can isolate a protein of interest by fusing its gene with that of an epitope tag, allowing the resulting fusion protein to be captured using an antibody that specifically recognizes the tag. This technique is called tag-based immunoprecipitation.
Immunoprecipitation
Immunoprecipitation is a common method used to isolate a particular protein from a mixture by using a specific antibody that binds to it. This process also captures any molecules—especially interacting proteins—that are attached to the target protein (Figure 1). The technique is especially useful for exploring protein–protein interactions. However, if a specific antibody for the target protein is not available or not efficient, researchers genetically fuse the gene of the target protein with a gene encoding a short, well-known epitope tag (like FLAG, HA, or Myc). When the tagged protein is expressed in cells, other proteins that bind to it precipitate with it. The identification of the associated proteins reveals some of the intracellular protein-protein interactions of the tagged protein.
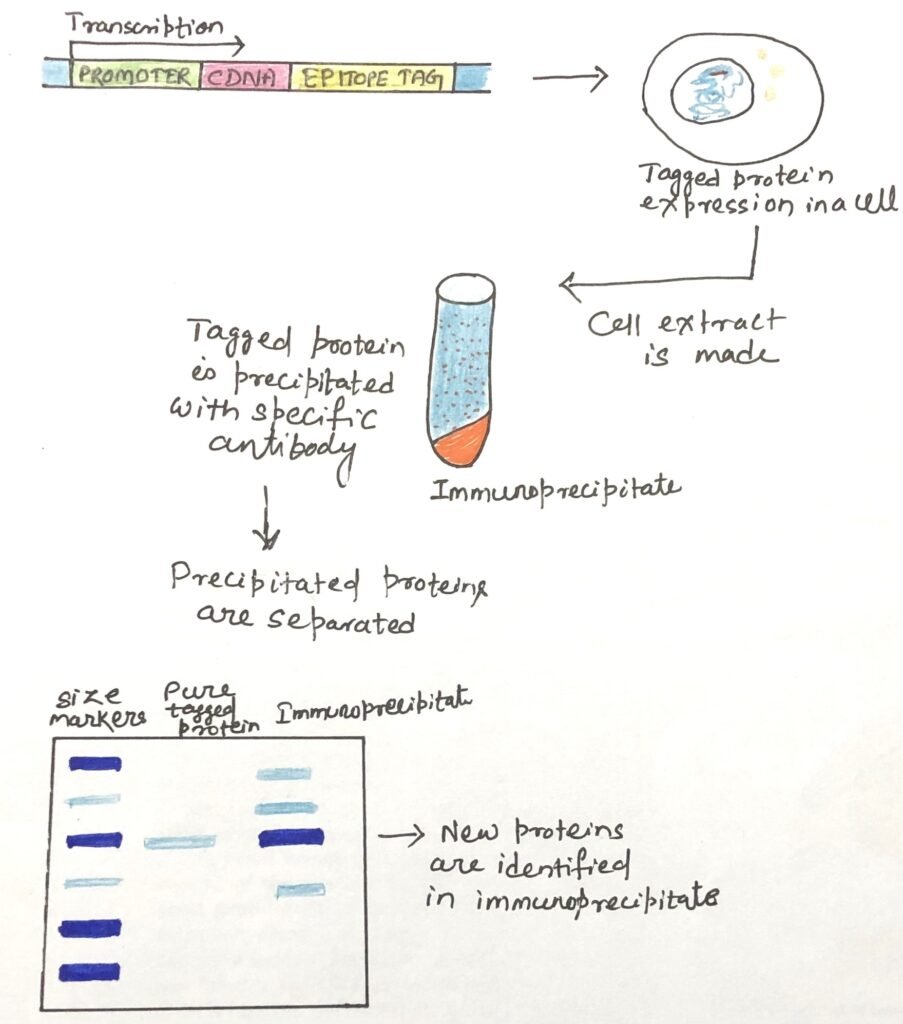
Affinity Chromatography for Identifying Protein-Protein Interactions
A crude cell extract containing a tagged protein is passed through a column with an immobilized antibody that binds specifically to the tag. As the tagged protein attaches to the antibody, any proteins that interact with it may also remain bound in the column. A specific protease is then used to cleave the tag from the protein, allowing the protein and its associated complexes to be eluted. This approach helps researchers map intricate networks of protein interactions within the cell. This chromatographic method for analyzing protein-protein interactions is versatile and can be applied using a range of protein tags (His tags, GST tags, FLAG tags, Myc tags, etc.) that can be immobilized on a suitable chromatographic medium.
Work of TAP Tags
Tandem affinity purification (TAP) tags improve the specificity of this chromatographic technique. In this method, the target protein is engineered to contain two sequential tags and is expressed within cells. The first tag, usually protein A (derived from the surface of Staphylococcus aureus), binds strongly to mammalian immunoglobulin G (IgG) (Figure 2). This tag binds tightly to mammalian immunoglobulin G (IgG). The second tag is a calmodulin-binding peptide, which is often used. A crude extract containing the TAP-tagged fusion protein is then passed through a column containing immobilized IgG antibodies, which specifically capture the protein A tag.
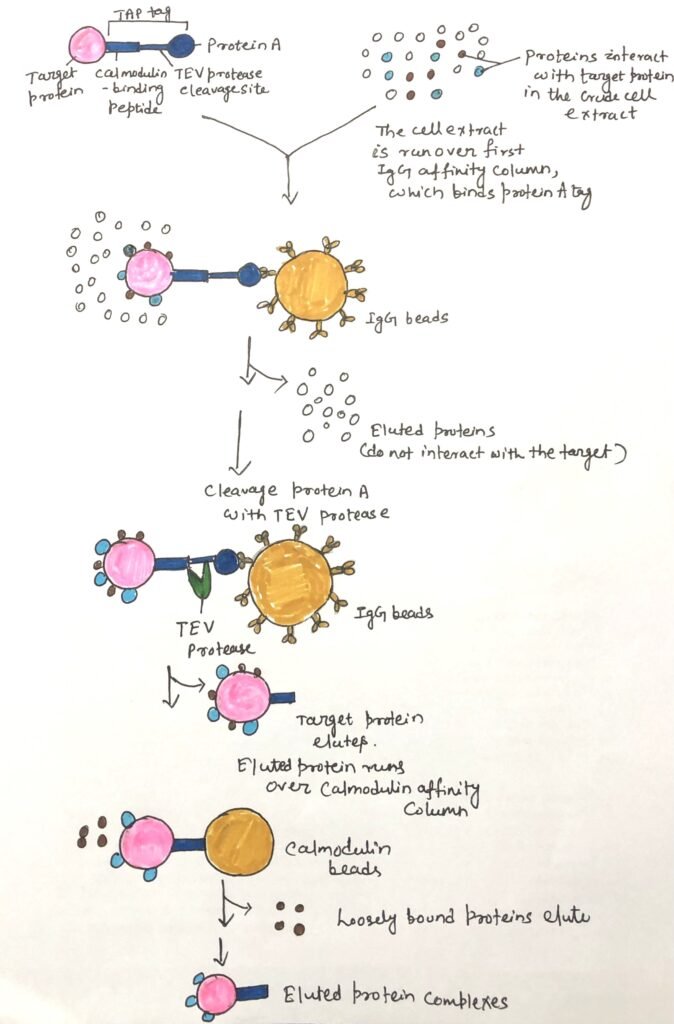
Most of the unbound cellular proteins are washed away during the column purification. However, proteins that interact with the target protein remain bound. The first tag is then enzymatically removed from the fusion protein. This allows the target protein and its non-covalently associated partners to be eluted. This eluate is subsequently passed through a second column containing a calmodulin-affinity matrix that captures the second tag.
Any weakly associated proteins are washed off. Finally, after cleaving the second tag, the target protein, along with its interacting partner, is eluted from the column. The two sequential purification steps help remove weakly bound contaminants. This reduces the chance of false positives. Thus, it ensures that the protein interactions remaining after both steps are more likely to be biologically meaningful.
Gal4 Protein Defines Protein-Protein Interaction
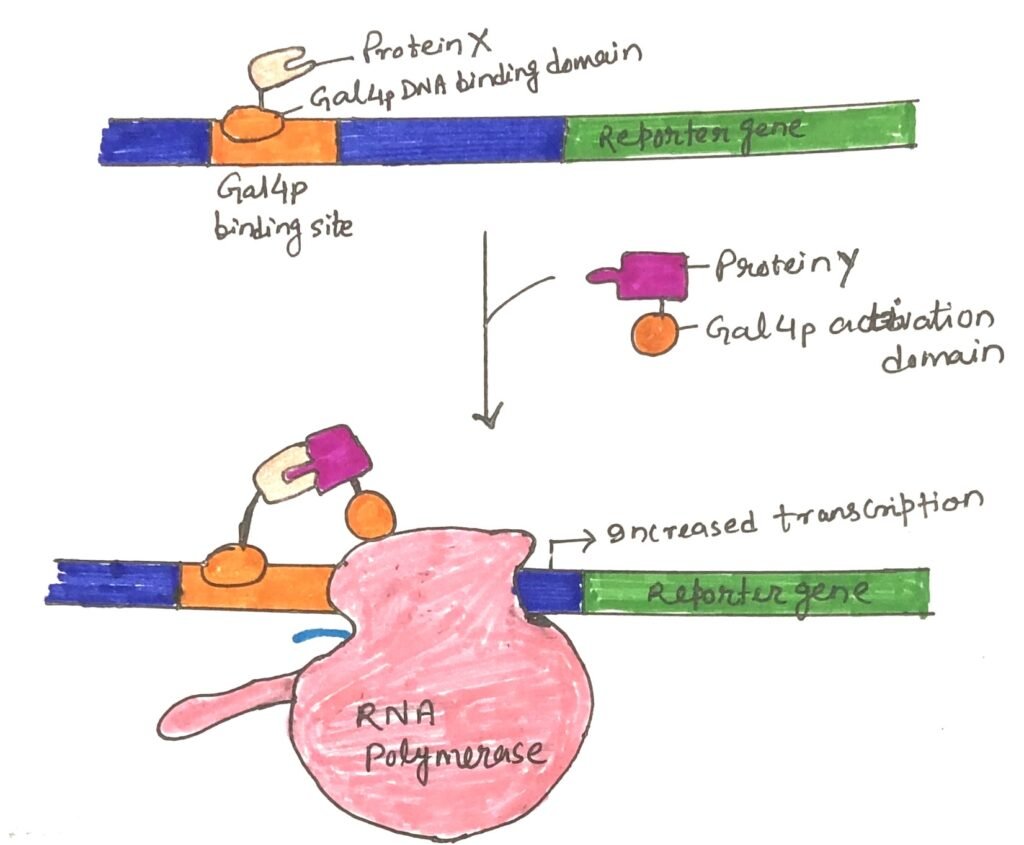
The Gal4 protein (Gal4p) helps define protein-protein interactions by serving as a reporter system in yeast two-hybrid assays. In yeast, Gal4p activates the transcription of GAL genes, which code for enzymes involved in galactose metabolism. Gal4p consists of two distinct domains—one that binds to a specific DNA sequence and another that activates RNA polymerase.
This activation enables the polymerase to transcribe mRNA from a nearby gene. Although these two domains are stable when separated, RNA polymerase activation only occurs when the activation domain is positioned correctly by the DNA-binding domain. Therefore, the two domains must come together to function effectively.
Yeast Two-Hybrid Assay
The coding sequences of the genes under investigation are linked to the yeast gene encoding either the DNA-binding domain or the activation domain of Gal4p. This results in the production of various fusion proteins. The activation of transcription is initiated when a protein fused to the DNA-binding domain interacts with a protein fused to the activation domain (Figure 3). This leads to the transcription of the reporter gene, which yields a protein. This protein either helps in growth or is required by an enzyme that catalyzes a reaction with a colored product.
A specific yeast strain can be used to create a library, where each cell carries a gene fused to the Gal4p DNA-binding domain. This library includes a wide range of different genes. A second yeast strain is engineered to contain a gene of interest fused to the Gal4p activation domain. When these two strains are mated, diploid cells form and grow into colonies (Figure 4). If the protein of interest interacts with a partner protein from the library, the interaction activates transcription of a reporter gene. It enables growth on selective media or the development of a detectable color.
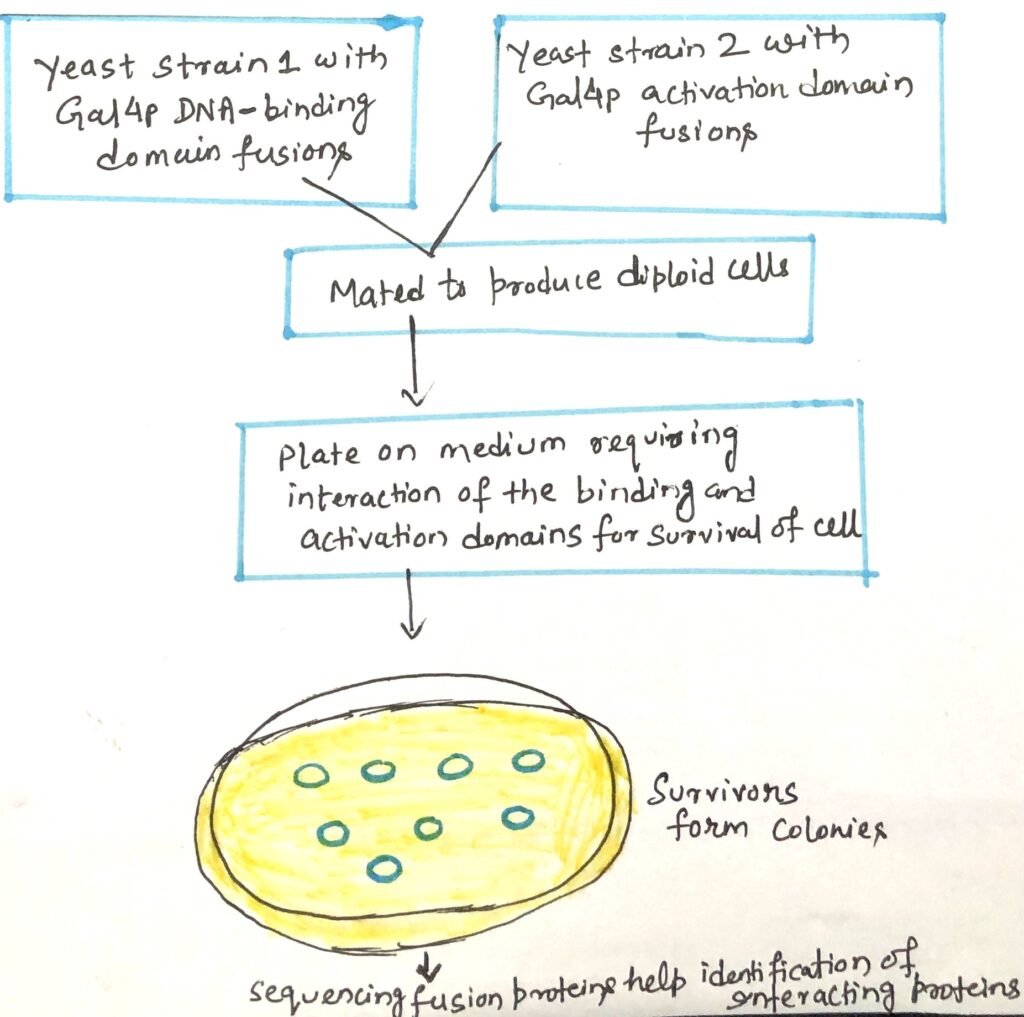
This system enables high-throughput screening to identify proteins that interact with the target protein. The interacting partner fused to the Gal4p DNA-binding domain in positive colonies can be rapidly identified by sequencing the corresponding fusion gene. However, the formation of multiprotein complexes may occasionally lead to false-positive results. The Techniques used to determine cellular localization and protein interactions provide important clues about a protein’s function.
Conclusion
The function of a specific protein can often be inferred by identifying the cellular components it binds to. Immunoprecipitation is a common method used to isolate a particular protein from a mixture by using a specific antibody that binds to it. This process also captures any molecules—especially interacting proteins—that are attached to the target protein.
Affinity chromatography helps analyze protein-protein interactions. It is versatile and can be applied using a range of protein tags (His tags, GST tags, FLAG tags, Myc tags, etc.) that can be immobilized on a suitable chromatographic medium. Tandem affinity purification (TAP) tags improve the specificity of this chromatographic technique. In this method, the target protein is engineered to contain two sequential tags and is expressed within cells.
The Gal4 protein (Gal4p) helps define protein-protein interactions by serving as a reporter system in yeast two-hybrid assays. In yeast, Gal4p activates the transcription of GAL genes, which code for enzymes involved in galactose metabolism. Gal4p consists of two distinct domains—one that binds to a specific DNA sequence and another that activates RNA polymerase. The two domains must come together to function effectively.
You may also like:
- Electrophoresis: The process of separation of proteins
- Green fluorescent protein helps identify proteins in cells

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.




