In this article, I briefly describe Southern blotting, which helps detect specific DNA sequences in a DNA sample.
Southern Blotting
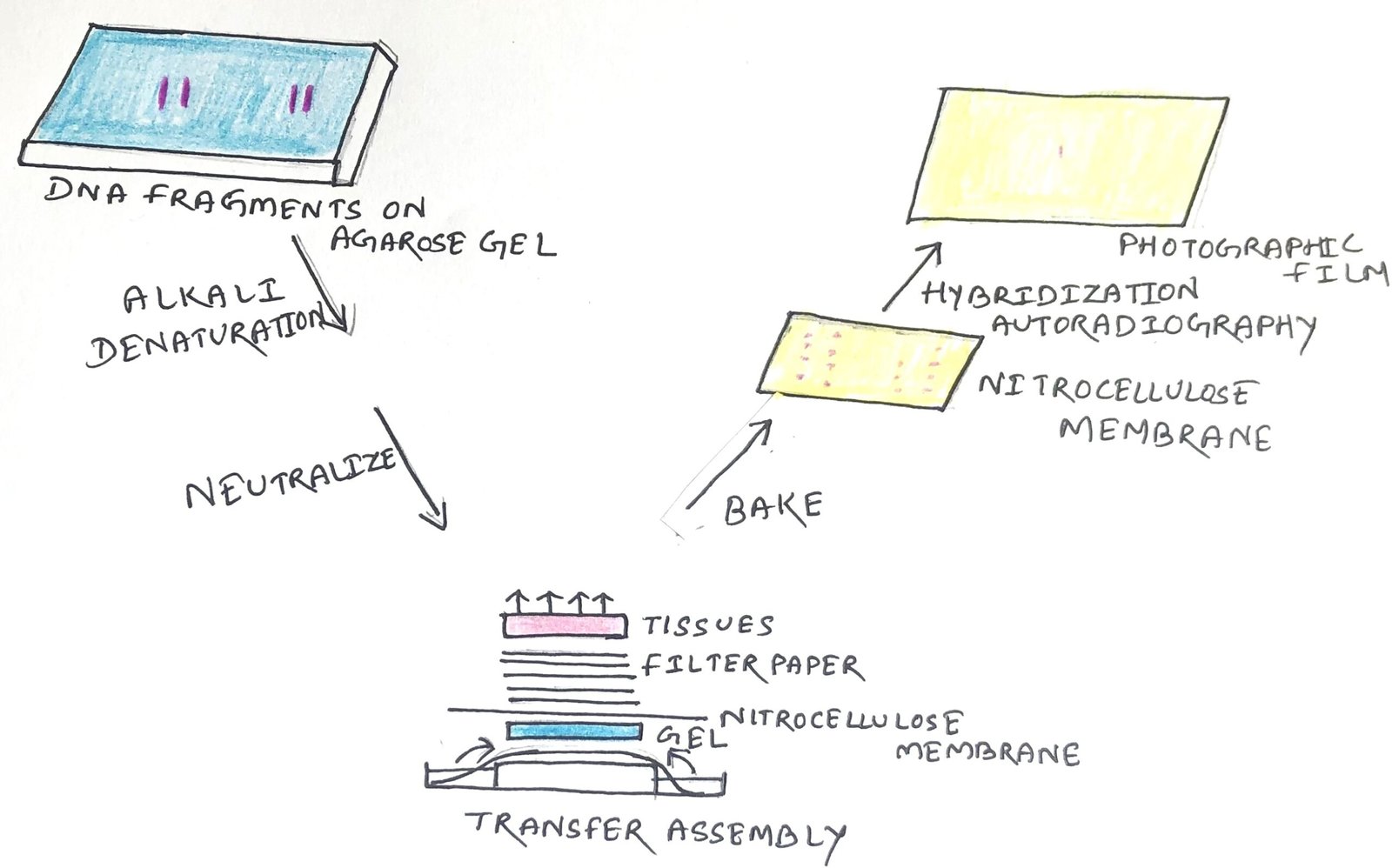
Southern blotting is a technique used in a laboratory to detect specific DNA sequences in a DNA sample. The method is named after its inventor, the British biologist Edwin Southern. The transfer of DNA fragments from an electrophoresis gel to a membrane support results in the immobilization of the DNA fragments. Thus, the membrane carries a semi-permanent reproduction of the banding pattern of the gel. The process of immobilization is followed by fragment detection through the probe hybridization technique.
Procedure
The procedure begins with restriction digestion of high molecular weight DNA strands into smaller fragments by restriction endonucleases. Then, the DNA fragments undergo agarose gel electrophoresis for size separation. The fragments are then transferred to a nitrocellulose membrane placed on top of the gel (Figure 1).
In Southern blotting, before DNA transfer, it is usually denatured with alkali. The denaturation in an alkaline environment may improve the binding of the negatively charged DNA to a positively charged membrane. It separates the DNA into single strands for later hybridization to the probe and destroys any residual RNA that may still be present in the DNA.

Blotting
A sheet of nitrocellulose membrane is placed on the top of the gel. The constant application of pressure to the gel ensures even contact between the gel and the membrane. There are five methods through which the transfer of the DNA fragments to the nitrocellulose membrane can be done.
Upward Capillary Transfer
In this method, the transfer of the DNA fragment is upward from the gel to the membrane, where the liquid or buffer flows upward.
Downward Capillary Transfer
In this method, the gel is placed on the surface of the nylon-charged membrane. Then the DNA fragments are transferred in a downward direction with the flow of the alkaline buffer.
Simultaneous Transfer to Two Membranes
Here, high concentrations of DNA fragments are transferred simultaneously from the gel to two membranes.
Electrophoretic Transfer
In this method, a large electric current makes the DNA transfer difficult due to the temperature of the buffer used. Thus, these machines can be used in cold areas or should be equipped with cooling machines.
Vacuum Transfer
In this process, a buffer transfers the DNA from the gel to the nitrocellulose or nylon membrane. The gel is placed directly on the membrane, and the membrane is placed on a porous screen in the vacuum chamber.
After the transfer process, the process of immobilization is started. Then, the membrane is baked in a vacuum or regular oven at 80 °C for 2 hours. It will help in the permanent attachment of the transferred DNA to the membrane.
Hybridization
The membrane is then exposed to a hybridization probe (a single DNA fragment with a specific sequence whose presence in the target DNA is to be determined). The probe DNA is labeled, so that it can be detected, usually by incorporating radioactivity or tagging the molecule with a fluorescent or chromogenic dye. After hybridization, an excess probe is washed from the membrane. Then, autoradiography helps to visualize the hybridization pattern on an X-ray film.
Analysis of Results
A probe that hybridizes only to a single DNA segment (which has not been cut by the restriction enzyme), will produce a single band on a Southern blot. Multiple bands will be observed when the probe hybridizes several highly similar sequences. The conditions of hybridization and washing are critical. If the probe and target are 100% identical in sequence, then a high stringency hybridization can be carried out. To improve specificity and reduce hybridization of the probe to sequences that are less than 100% identical, the hybridization parameters may be changed.
The hybridization temperature and the salt concentration in the hybridization buffer determine the stringency. For probes that don’t match the target completely, the stringency must be reduced to a level that allows imperfect hybrids to form. If the stringency of the hybridization is too low, then the probe may bind to too many sequences to be useful. The involvement of formamide in the hybridization buffer reduces the actual hybridization temperature by about 25°C, i.e., from the usual 68°C to the more convenient 43°C.
Applications of Southern Blotting
Southern hybridization technique can be applied to locate the exact position of a cloned gene within a recombinant DNA molecule. The cloned DNA fragment is relatively large, whereas the gene of interest is less than 1kb in size, contained somewhere in the cloned fragment.
Southern blots of cloned genomic DNA fragments can be probed with cDNA to find the corresponding part of the genomic clone to the cDNA fragment. If the Southern blot contains genomic DNA fragments from the whole genome, the probe will give information about the size of the gene fragment on the genome and the number of copies of the gene in the genome.
Southern blotting helps to study normal chromosomal or gene rearrangement. Genes change due to insertions, deletions, rearrangements, and point mutations. Southern hybridization aids in locating these changes in genes.
It is used to determine the altered recognition site due to a single-nucleotide polymorphism, which changes a specific restriction enzyme. Southern blotting helps identify methylated sites in particular genes. It can be applied to personal identification through fingerprinting and in disease diagnosis.
However, the limitations of Southern blotting lie in the complexity of the process. This is a multi-step process that requires expensive equipment and reagents. It requires high-quality DNA in large quantities. It is a time-consuming method and can’t detect mutations at the base-pair level.
Conclusion
Southern blotting is a laboratory technique invented by the British biologist Edwin Southern. It helps to detect a specific DNA sequence in a DNA sample. DNA fragments are transferred from an electrophoresis gel to a membrane support that resulting in the immobilization of the DNA fragments.
Southern blotting begins with restriction digestion of high molecular weight DNA strands into smaller fragments. After digestion, the DNA fragments undergo agarose gel electrophoresis for size separation. Then, the fragments are transferred to a nitrocellulose membrane placed on top of the gel.
A sheet of nitrocellulose membrane is placed on top of the gel. The constant application of pressure to the gel ensures even contact between the gel and the membrane. The five methods through which the transfer of the DNA fragments to the nitrocellulose membrane can be done are upward capillary transfer, downward capillary transfer, electrophoretic transfer, simultaneous transfer to two membranes, and vacuum transfer.
The membrane is then exposed to a hybridization probe (a single DNA fragment with a specific sequence whose presence in the target DNA is to be determined). The probe DNA is labeled so that it can be detected. Then, with a radioactive or fluorescent probe, the hybridization pattern can be seen on an X-ray film by autoradiography.
Southern hybridization technique can be applied to locate the exact position of a cloned gene within a recombinant DNA molecule. It helps to study the normal chromosomal or gene rearrangement. Southern blotting helps identify methylated sites in particular genes. However, Southern blotting is a complex, time-consuming process that requires expensive equipment and reagents. It also requires high-quality DNA in large quantities.
You may also like:
- Northern blotting: Detection of RNA
- Western blotting: Identifying a specific protein
- DNA Beyond the Double Helix: Structural Variants and Unusual Sequences
- Advanced DNA Sequencing Technologies

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.