In this article, I briefly describe the organization of cloned genes.
cDNA clones
The cDNA clones synthesized using oligo(dT) as a primer, have a defined organization. The exact copy of an mRNA molecule is called a cDNA or complementary DNA. The well-characterized cDNA molecule binds with a befitting vector and the combination (vector containing the cDNA) is introduced into a bacterium. The transformed bacterial cell containing a plasmid with a DNA copy of an mRNA molecule is known as a cDNA clone. It isn’t easy to obtain cDNA from double-stranded DNA molecules. Thus, most cDNA clones have been prepared from mRNA sequences from eukaryotic cells.
Usually, the 3’ end of the clone consists of a run of A residues. At some variable distance upstream, there will be an open reading frame (ORF) ending in a stop codon. Genomic clones from eukaryotes are larger, and may also contain intron sequences and non-transcribed sequences. Thus, it’s a great challenge to understand their organization. The genomic sequences upstream of the transcription start site and downstream of the 3’ processing site along with introns are usually absent in the cDNA clones.
The procedures to locate the 5’ end of an RNA transcript
S1 nuclease mapping
It is a laboratory technique, used to locate the precise 5’ end and 3’ end of an RNA transcript. The S1 nuclease is an endonuclease that can degrade single-stranded DNA and RNA. The enzyme is used to cut protruding single-stranded termini from double-stranded DNA.
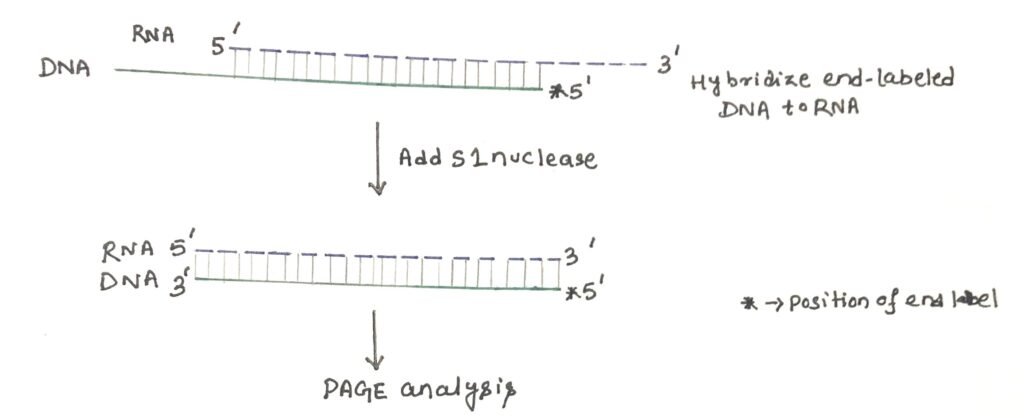
For 5′ end mapping, an end-labeled antisense DNA molecule is hybridized to the RNA preparation (figure 1). The hybrids are then treated with the single-strand specific S1 nuclease, which will remove the single-strand protrusions at each end. The remaining material is analyzed by polyacrylamide gel electrophoresis next to size markers or a sequencing ladder. By applying the process of autoradiography, the size of the nuclease-resistant band is revealed. This allows the end of the RNA molecule to be deduced.
Primer extension
The 5’ ends of RNA or DNA can be mapped by the technique of primer extension. For a known gene, the start site of RNA transcription can be determined by this technique. In this method, a radiolabelled primer, of length 20-50 nucleotides is required, which is complementary to a region near the 5’ end of the gene (figure 2).
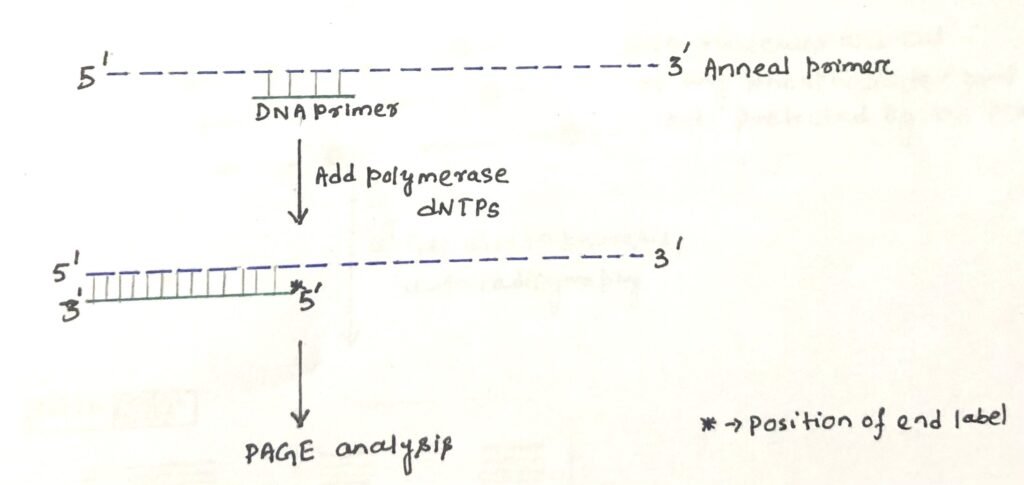
The primer is annealed to the RNA and the enzyme reverse transcriptase is used to synthesize cDNA until it reaches the 5′ end of the RNA. It is possible to determine the transcriptional start site by running the product on a polyacrylamide gel. The length of the sequence on the gel represents the distance from the start site to the radiolabelled primer.
Studying protein-nucleic acid interactions
Gel retardation assay
It is a rapid and sensitive technique used to study protein-nucleic acid interactions. In this assay, solutions of protein and nucleic acid are combined and the resulting mixtures are subjected to electrophoresis under native conditions through polyacrylamide or agarose gel. After electrophoresis, the distribution of species containing nucleic acid is determined by autoradiography. It shows the effect of protein binding to a labeled nucleic acid and can be used to detect transcription factors binding to regulatory sequences (figure 3).
A cell or a nuclear extract expected to contain the binding protein is mixed with a short labeled nucleic acid ( the region of a genomic clone upstream of the transcription start site). Samples of labeled nucleic acid, with and without extract, run on a non-denaturing gel (agarose or polyacrylamide). If a large excess of unlabeled nucleic acid of different sequences is present, it will bind proteins that interact nonspecifically. Then, the specific binding of a factor to the labeled molecule to form one or more DNA -protein complexes is shown by the presence of slowly migrating (retarded) bands on the gel by autoradiography. In general, protein-nucleic acid complexes migrate more slowly than the corresponding free nucleic acid.
Gel retardation shows the binding of a protein to a DNA molecule. However, it doesn’t provide the sequence of the binding site which could be anywhere in the fragment used. DNase footprinting shows the actual region of sequence with which the protein interacts.
DNase I footprinting
It is a DNA footprinting technique that detects DNA-protein interaction. The binding protein often protects the DNA from enzymatic cleavage, which makes it possible to locate a protein binding site on a particular DNA molecule. This technique involves an enzyme, deoxyribonuclease (DNase I). The enzyme deoxyribonuclease cuts the radioactively end-labeled DNA, followed by gel electrophoresis to detect the cleavage pattern.
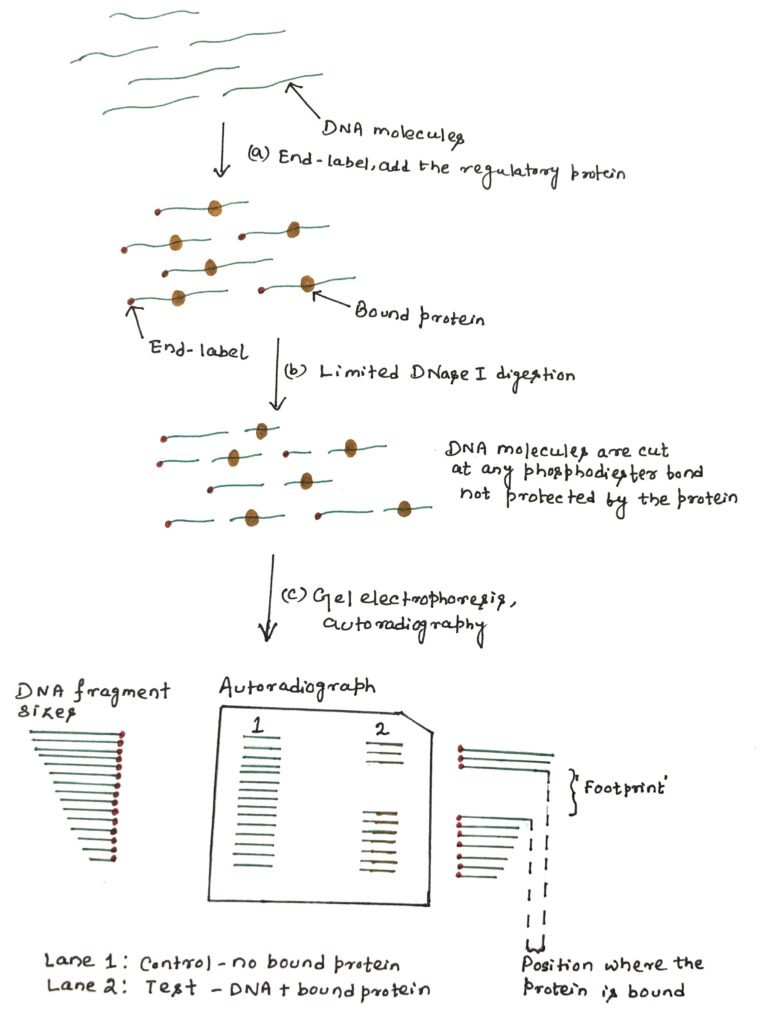
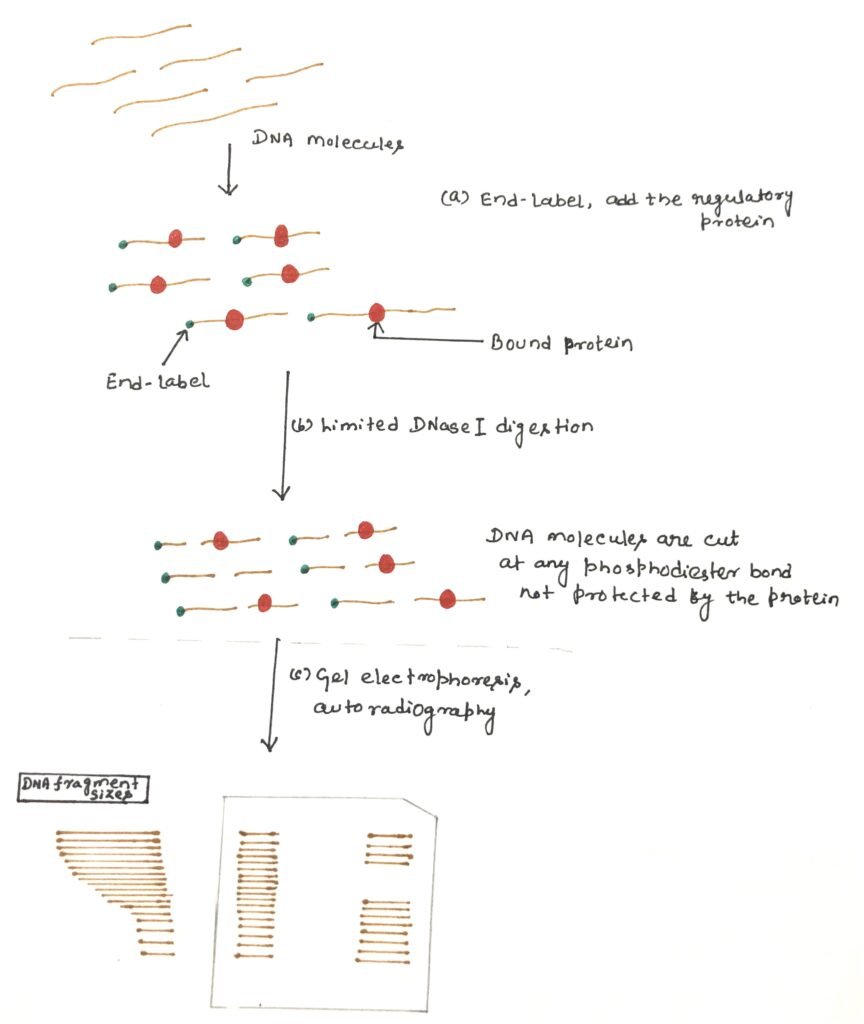
An end-labeled DNA fragment is mixed with the protein preparation. After binding, the complex is gently digested with DNase I to produce one cleavage per molecule on average. In the region where the protein binds, the nuclease can’t easily gain access to the DNA backbone, thus creating a few cuts (figure 4). When the partially digested DNA is analyzed by polyacrylamide gel electrophoresis (PAGE), a ladder of bands is seen.
These bands show all the random nuclease cleavage positions in control DNA. These cleavage patterns of DNA in the absence of a DNA-binding protein, are known as free DNA. In the lane, where protein was added, the ladder will have a gap, or region of reduced cleavage, corresponding to the protein-binding site where the protein has protected the DNA from nuclease digestion. This protection will result in a clear area on the gel, which is referred to as the ‘footprint’. The binding affinity of the protein can be estimated according to the minimum concentration of protein at which a footprint is observed.
Conclusion
The cDNA clones synthesized using oligo(dT) as a primer, have a defined organization. It is difficult to obtain cDNA from double-stranded DNA molecules. Thus, most cDNA clones have been prepared from mRNA sequences from eukaryotic cells. Genomic clones from eukaryotes are larger, and may also contain intron sequences, as well as non-transcribed sequences. Thus, making it an onerous task to understand their organization.
S1 nuclease mapping determines the precise 5′- and 3′- ends of RNA transcripts. The RNA-DNA hybrids are treated with the single-strand specific S1 nuclease, which will remove the single-strand protrusions at each end. Then it undergoes polyacrylamide gel electrophoresis (PAGE).
Primer extension is a technique whereby the 5′ ends of RNA or DNA can be mapped. The primer extension can be used to determine the start site of RNA transcription for a known gene. This technique requires a radiolabelled primer (usually 20 – 50 nucleotides in length) which is complementary to a region near the 5′ end of the gene.
Gel retardation is a technique, which shows the effect of protein binding to a labeled nucleic acid and can be used to detect transcription factors binding to regulatory sequences.
Gel retardation shows the DNA-protein binding, but it doesn’t provide the sequence of the binding site, which could be anywhere in the fragment used. DNase footprinting shows the actual region of sequence with which the protein interacts. The binding protein often protects the DNA from enzymatic cleavage, which makes it possible to locate a protein binding site on a particular DNA molecule. This technique involves an enzyme, deoxyribonuclease (DNase I).
You may also like:

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.



