In this article, I briefly describe the methods of clone identification.
Gene library
The collection of different DNA sequences from an organism, which is the total genomic DNA of the organism, makes a gene library. Each DNA sequence is cloned into a vector for ease of purification, storage, and analysis. The first step in the construction of a gene library is to extract the organism’s DNA from cells. After the DNA is extracted, it is followed by digestion with a restriction enzyme to cut it into fragments of a specific size. Then, these fragments are inserted into a vector by the enzyme DNA ligase. For easy amplification and analysis of specific clones from the library, the vector is taken up by a host organism like a population of E.coli or yeast.
Depending upon the source of DNA, two types of gene libraries can be prepared. If the DNA is genomic, the library is called a genomic library. If the DNA is a copy of an mRNA population, which is cDNA, then the library is a cDNA library. A library is called representative when the starting material is well represented by it, and the library contains all the original sequences.
Once a suitable library has been prepared, several procedures can be employed to attempt the identification of the desired clone. Hybridization probing is one of the important techniques for clone identification.
Colony and plaque hybridization
It is a method of selection of bacterial colonies with desired genes through cloning and transfer. Colony hybridization begins with the extraction of a DNA segment containing a specific gene and inserting it into a bacteria plasmid through recombination. These bacterial plasmids are cultured on a nutrient agar plate, leading to the production of bacteria colonies. In 1975, Grunstein and Hogness developed a screening procedure to detect DNA sequences in transformed bacterial colonies or bacteriophage plaques by hybridization in situ with a radioactive probe. Depending on the probe specificity, this can lead to the rapid identification of one colony among thousands of colonies.
The colonies to be screened are first symmetrically replicated onto a nitrocellulose or nylon membrane. A replica agar plate is retained as a reference set. The colonies are grown, and the nitrocellulose membrane disc carrying the colonies is removed. Then, it is treated with an alkali to lyse the bacterial colonies and denature their DNAs (figure 1). The membrane disc is then treated with proteinase K to remove proteins and leave the denatured DNA bound to the nitrocellulose membrane. The membrane undergoes a short heat treatment at 80°C or UV radiation so that the single-stranded DNA molecules can bind tightly to it. The molecules will be attached to the membrane through their sugar-phosphate backbones. Thus, the base will be free to pair with complementary nucleic acid molecules.
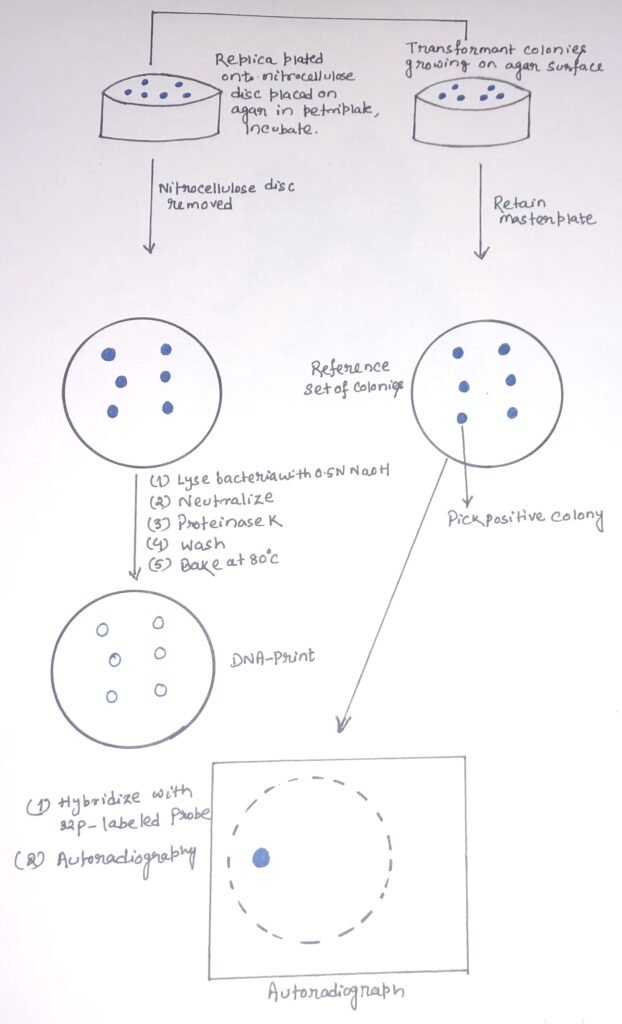
Nucleic acid hybridization and autoradiography
The probe must be labeled and denatured by heating. Then, it is applied to the membrane in a solution of chemicals that promote nucleic acid hybridization. After a short period of hybridization, the membrane is washed to remove any unbound probe molecules. Then, it is dried and the positions of the bound probe are detected. During the process of hybridization, any colony carrying a sequence complementary to the probe will become radio-labeled. Then, it can be identified by autoradiography. A clone showing a positive result can be picked from the master plate and grown to provide DNA for further analysis.
Immunological screening
It is a process where small amounts of proteins made in a bacterial cell can be detected. This method not only allows the detection of a complete protein but also a fragment of a protein sequence by its antigenic determinants. Immunological detection of clones synthesizing a foreign protein has also been successful where the inserted gene sequence is expressed.
In this method, transformed cells are plated on agar. The colonies are killed by subsequent procedures. So, the preparation of a replica plate is necessary. The bacterial colonies are then lysed to release the antigen from positive colonies. Nitrocellulose filters imprinted with detritus of bacterial lysis are soaked in a solution containing the unlabeled antibody. The antigen forms complexes with the bound IgG antibody. The filter is removed and exposed to125I labeled IgG antibody. The 125I-IgG can react with the bound antigen via antigenic determinants at sites other than those involved in the initial binding of antigen to the IgG-coated filter. The filter is washed, and in an autoradiographic image, positively reacting colonies are detected. The required clones can be recovered from the replica plate.
Chromosome walking
It is a technique used to isolate and clone a particular gene or allele through positional cloning. It is widely used for the discovery of new genes and chromosome sequencing. The process is used to move systematically along a chromosome from a known location and to clone overlapping genomic clones that represent progressively longer parts of a particular chromosome (figure 2).
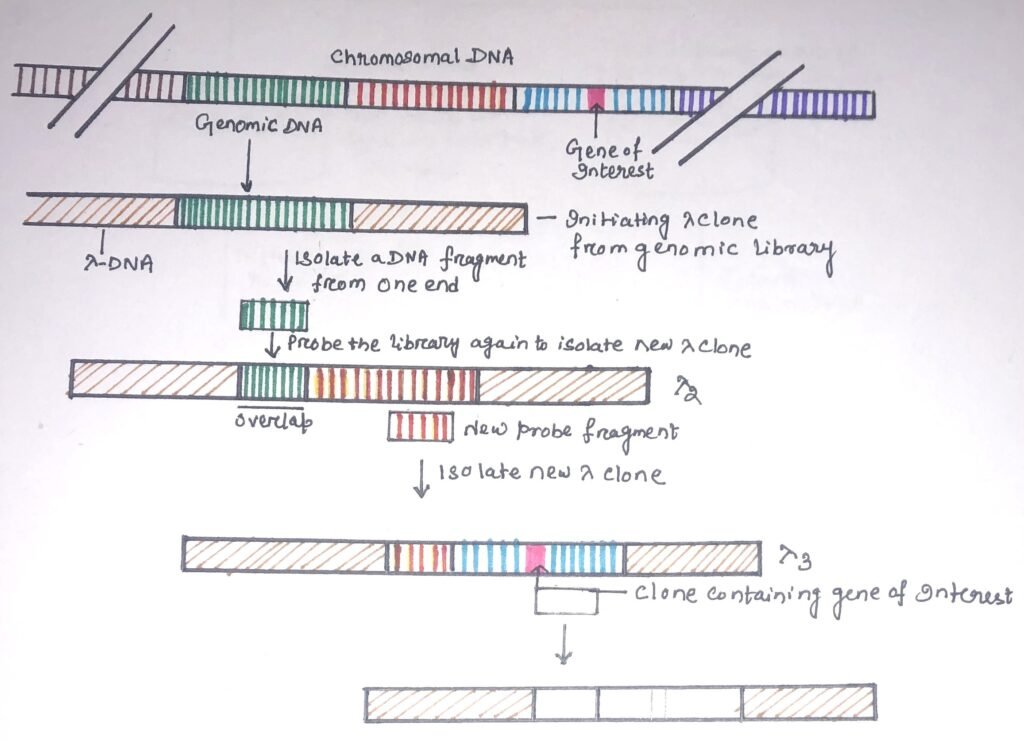
A small segment of DNA from one end of the genomic clone is used as a probe to isolate clones containing this sequence and adjacent sequences encoding the next portion of the genome. The end sequence of the second clone is used to isolate a third clone and so forth until a series of overlapping clones are isolated. DNA probes with single-copy sequences are used because a repeated sequence probe can produce several unrelated recombinants.
If the gene library does not contain a suitable sequence, it may lead to the breakdown of chromosome walk. In this situation, a technique called chromosome jumping can be used. Chromosome jumping can also be used when the ‘walk’ distance is quite large, e.g., positional cloning from a relatively distant start point. When DNA is circularized or cloned into a cosmid vector, the opposite ends of the long DNA fragment come together. Without mapping the entire DNA, the end fragments can be used as successive probes to screen a library. This can allow the isolation of clones that are around 50kb from each other, so producing a more rapid ‘walk’.
Conclusion
After the preparation of a suitable gene library, several procedures can be employed to attempt the identification of the desired clone. Hybridization probing is one of the important techniques for clone identification.
Colony and plaque hybridization is a method of selection of bacterial colonies with desired genes through cloning and transfer. Colony hybridization begins with the extraction of a DNA segment containing a specific gene and inserting it into a bacteria plasmid through recombination. In 1975, Grunstein and Hogness developed a screening procedure to detect DNA sequences in transformed bacterial colonies or bacteriophage plaques by hybridization in situ with a radioactive probe.
Small amounts of proteins made in a bacterial cell can be detected through immunological screening. Chromosome walking is a technique used to isolate and clone a particular gene or allele through positional cloning. It is widely used for the discovery of new genes and chromosome sequencing.
you may also like:

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around twelve years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.
