In this article, I briefly describe how green fluorescent protein helps identify proteins in cells.
Protein Localization and Function
Locating a gene product within the cell helps provide significant insight into its function. For instance, a protein found solely in the nucleus is likely to participate in nuclear-specific processes such as transcription, DNA replication, or chromatin organization. To pinpoint the location of proteins, researchers commonly create fusion proteins. These are engineered by attaching marker proteins that help visualize the protein’s position either directly or through techniques like immunofluorescence.
Green Fluorescent Protein (GFP)
This protein is derived from the jellyfish Aequorea Victoria. The green fluorescent protein (GFP) emits green light when illuminated with blue to ultraviolet light. This protein has also been identified in various other organisms such as corals, sea anemones, zoanthids, copepods, and lancelets. The protein has a β-barrel structure with a fluorophore (Figure 1), the fluorescent component of the protein in the center.
Autocatalytic Nature of GFP Fluorophore Formation
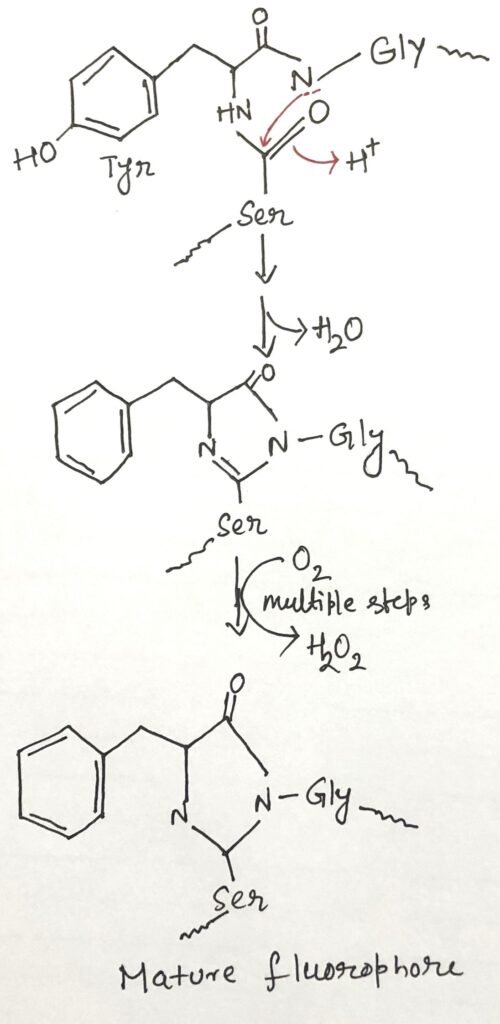
The fluorescent property of GFP arises from the rearrangement and oxidation of three specific amino acids (figure 1) within the protein. This process is autocatalytic, meaning it occurs without the need for additional proteins or cofactors—only molecular oxygen is required. As a result, GFP can be easily expressed in its active form in nearly any cell type. Remarkably, even a small number of GFP molecules is detectable under a microscope. It enables detailed observation of their location and movement within cells.
Roger Tsien carefully did protein engineering coupled with the isolation of related fluorescent proteins from other marine coelenterates. He has made variants of these proteins available in an array of colors and different characteristics, such as brightness and stability. If attaching GFP to a protein doesn’t affect its function or characteristics, the resulting fusion protein can be used to track where the protein is located in the cell under various conditions and to observe its interactions with other tagged proteins. For instance, the protein GLR1, a glutamate receptor of nervous tissue, has been visualized as a GLR1-GFP fusion protein in the nematode Caenorhabditis elegans.
Immunofluorescence as an Alternative for Protein Localization
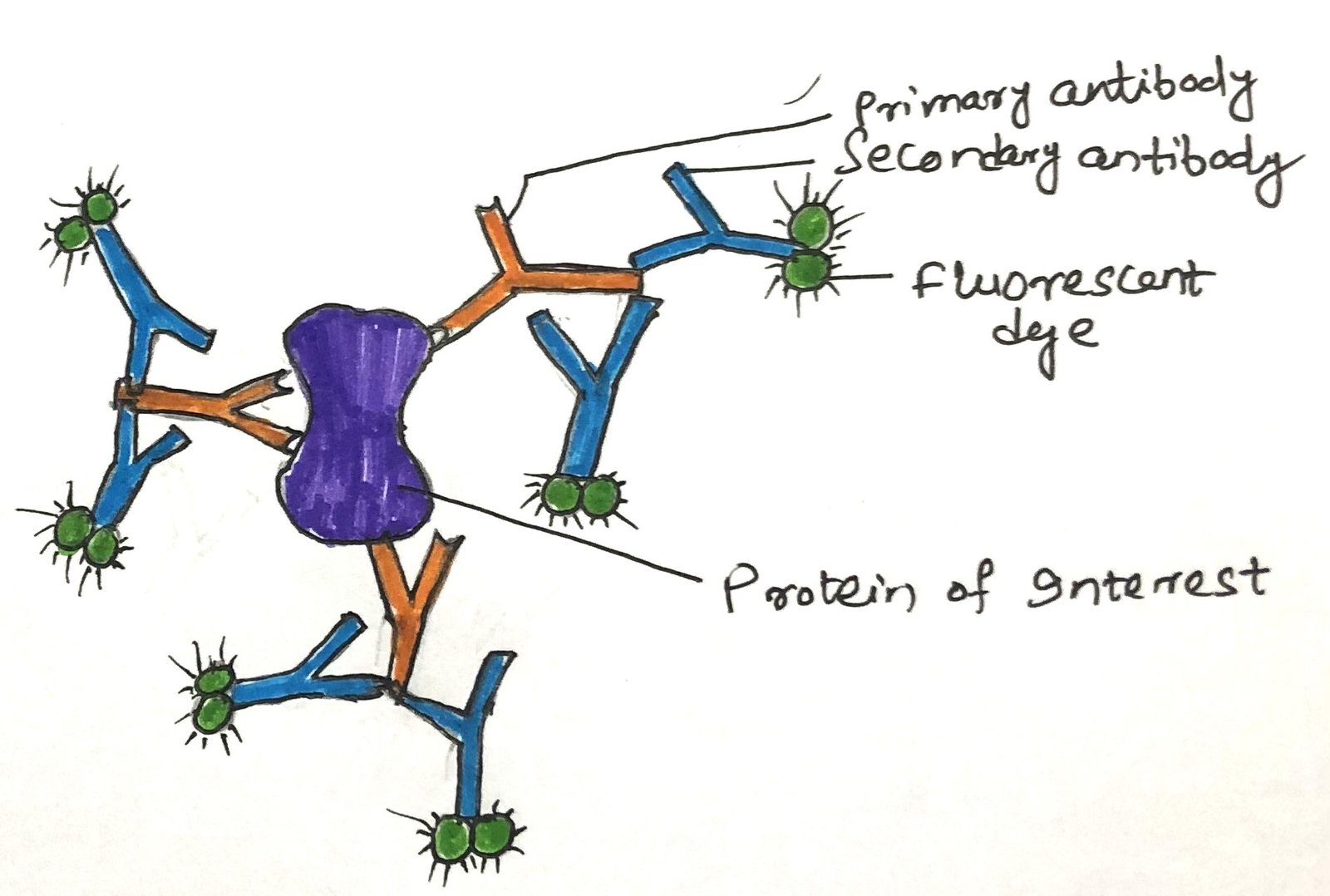
In a living cell, the visualization of a GFP fusion protein is not possible or even desirable. When a GFP fusion protein is either nonfunctional or not produced in high enough amounts for detection, immunofluorescence offers a reliable alternative for visualizing the native protein. This method involves fixing the cells and using antibodies to detect the protein of interest. Sometimes, the protein is engineered with an epitope tag—a short amino acid sequence recognized by a specific, widely available antibody to which fluorescent markers are attached. However, more often, the unmodified protein is directly targeted using an antibody that specifically binds to it. After this step, a second antibody is added that specifically binds to the first one. It is the second antibody with the attached fluorochrome(s) (Figure 2).
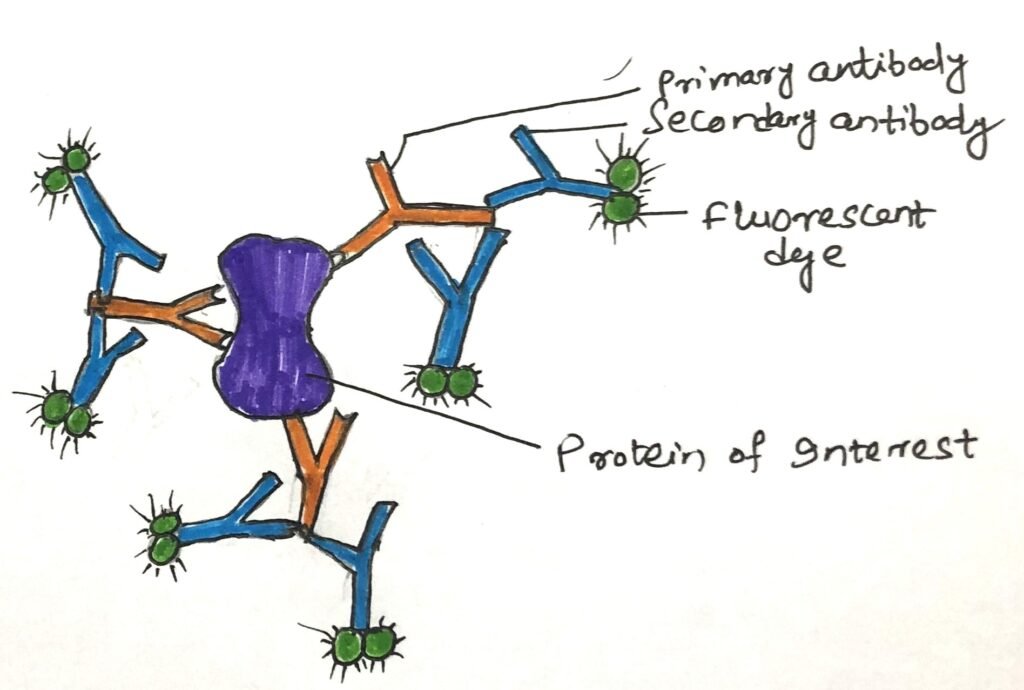
In an alternative version of this indirect detection method, the primary antibody is tagged with biotin, and a streptavidin-fluorochrome complex is applied afterward. The exceptionally strong and specific interaction between biotin and streptavidin allows for the attachment of multiple fluorophores. It greatly enhances the sensitivity of detection. Ultimately, these techniques enable the visualization of proteins in cells under a microscope. In a microscope, their presence is indicated by distinct points of fluorescence.
Visualizing Gene Expression with Reporter Constructs
Highly specialized cDNA libraries can be created by inserting cDNA sequences or fragments into a vector that links each one to a marker gene known as a reporter gene. This combination forms what’s called a reporter construct. A common example is fusing each gene in the library to the GFP (Green Fluorescent Protein) gene. As a result, each cell in the library expresses a unique fusion gene. If the fusion protein maintains its typical function and proper localization, the location of the gene product can be visualized as glowing spots within cells that express the gene at detectable levels.
Conclusion
Green fluorescent protein (GFP) is derived from the jellyfish Aequorea Victoria. The GFP emits green light when illuminated with blue to ultraviolet light. The protein has a β-barrel structure with a fluorophore, the fluorescent component of the protein, in the center.
The fluorescent property of GFP arises from the rearrangement and oxidation of three specific amino acids within the protein. Remarkably, even a small number of GFP molecules are detectable under a microscope, enabling detailed observation of their location and movement within cells.
When a GFP fusion protein is either nonfunctional or not produced in high enough amounts for detection, immunofluorescence offers a reliable alternative for visualizing the native protein. This method involves fixing the cells and using antibodies to detect the protein of interest.
Highly specialized cDNA libraries can be made by inserting cDNA fragments into vectors containing a reporter gene like GFP. This creates reporter constructs, where each cell expresses a unique fusion protein. If the fusion retains normal function and localization, the gene product’s location can be seen as glowing spots in cells with detectable expression.
You may also like:
- Western blotting: Identifying a specific protein
- Expression of recombinant proteins
- Deciphering Protein Function through Analysis of Protein–Protein Interactions
- Conventional Imaging Techniques Based on Immunofluorescence

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.