In this article, I briefly describe Northern blotting and its applications.
Northern blotting
It is a laboratory technique, which studies gene expression by detecting a specific RNA sequence in a blood or tissue sample. Northern blotting was first developed in 1977 by James Alwine, David Kemp, and George Stark at Stanford University. The name was extrapolated from Southern blotting named for biologist Edwin Southern. The process involves the size-separation of RNA samples by electrophoresis. Then, fragment detection is done through probe hybridization.
The procedure
The Northern blotting procedure begins with, the extraction of total RNA from a homogenized tissue sample or cells. The RNA molecules are separated by agarose gel electrophoresis and then transferred to a nitrocellulose membrane (figure 1). However, in Northern blotting, alkali denaturation is unnecessary for RNA and would in any case hydrolyze the molecules.
Northern blotting uses a nylon membrane with a positive charge as the negatively charged nucleic acids have a high affinity for them. The separated RNA sequence is transferred to the nylon membrane by two methods, such as capillary action and ionic interaction.
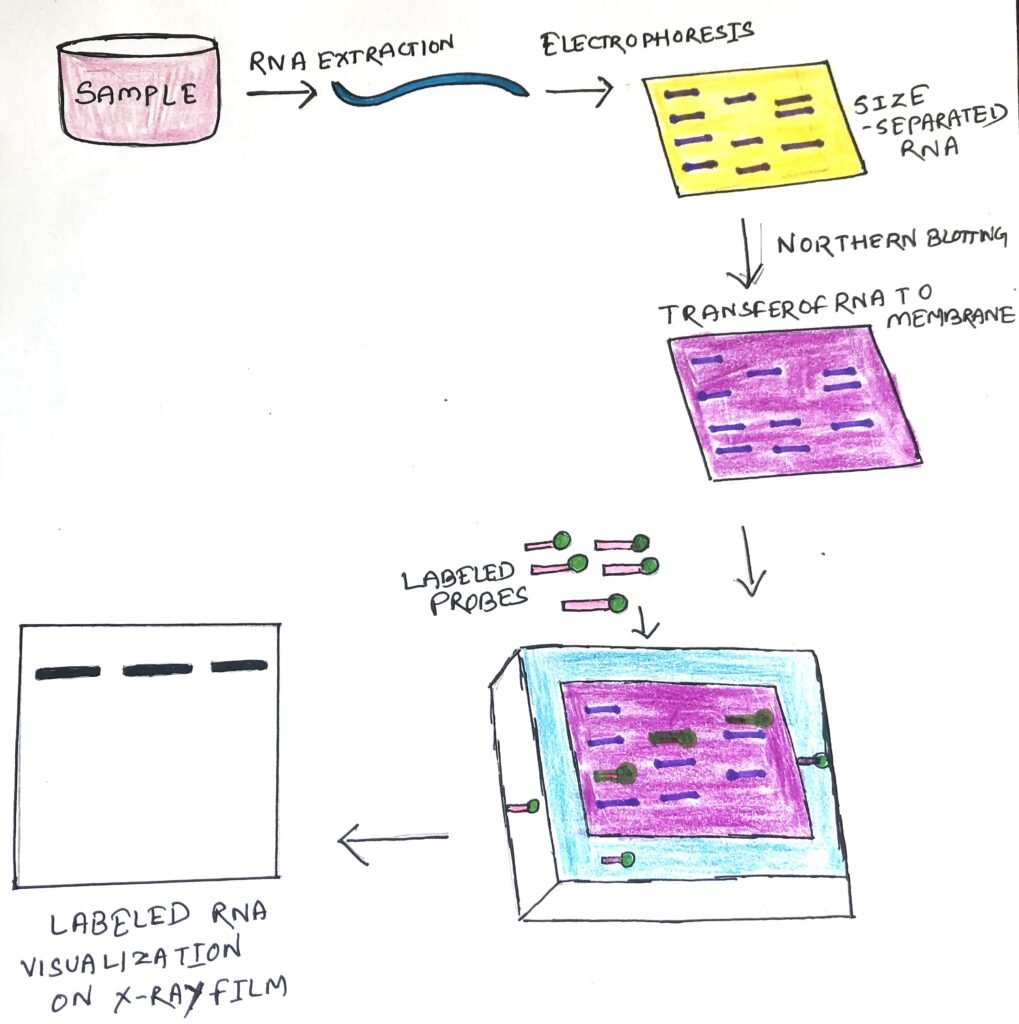
The transfer procedure begins with placing the agarose buffer on the bottom of the stack, followed by the blotting membrane. A small weight is placed on top of these paper towels. The entire setup is placed in a container containing the transfer buffer. At high temperatures, RNA degradation is prevented by formamide, in the transfer buffer. Formamide lowers the annealing temperature of the probe-RNA interaction, thus preventing RNA degradation.
Once the RNA has been transferred to the membrane, it is immobilized through covalent linkage, to the membrane by UV light or heat. A probe is labeled (figure 1) and is hybridized to the RNA on the membrane. Some experimental conditions can affect the efficiency and specificity of hybridization. These include ionic strength, viscosity, duplex length, mismatched base pairs, and base composition. The blotting membrane is washed to remove any unwanted probes. The hybrid signals are then detected by X-ray film.
Gels and probes used in Northern blotting
Generally, agarose gels are used to separate the RNA samples. Agarose gels contain formaldehyde as a denaturing agent for the RNA to limit secondary structure. However, polyacrylamide gel electrophoresis with urea (a denaturing agent) can be used for small and micro RNA sequences. Ethidium bromide can be used as a staining agent for gels. The size of the RNA fragments can be known by running an RNA ladder alongside the samples.
Probes used in Northern blotting can be RNA, DNA, or oligonucleotides of 25 complementary base pairs to the target RNA. They can be complimentary to, the whole RNA or part of the RNA of interest. For the RNA sequence of interest, cDNA is created with labeled primers to act as a probe in the Northern blot. The probes must be labeled with either radioactive isotopes or chemiluminescence. The chemiluminescent labeling can be done by attaching the probe to the enzyme or the probe is labeled with a ligand.
Applications of Northern blotting
Northern blotting helps to analyze a particular gene’s expression levels during morphogenesis and differentiation. It shows the gene’s expression at different environmental stress levels, during a pathogen infection and throughout treatment. Thus, it is possible to observe cellular control over structure and function.
Generally, the analysis of gene expression is done by microarrays. However, Northern blot analysis can detect small changes in gene expression that microarrays can’t. Microarrays can visualize thousands of genes at a time, whereas Northern blot can do it for, a small number of genes.
Northern blot gives information about the size of the mRNA and any precursors. It helps to identify any transcribed regions in a genomic clone.
Conclusion
Northern blotting is a laboratory technique, which studies gene expression by detecting a specific RNA sequence in a blood or tissue sample. It was first developed in 1977 by James Alwine, David Kemp, and George Stark at Stanford University.
The procedure begins with, the extraction of total RNA from a homogenized tissue sample or cells. The RNA molecules are separated by agarose gel electrophoresis, followed by their transfer to a nitrocellulose membrane. The transfer procedure begins with placing the agarose buffer on the bottom of the stack, followed by the blotting membrane.
Once the RNA has been transferred to the membrane, it is immobilized through covalent linkage, to the membrane by UV light or heat. A probe is labeled and is hybridized to the RNA on the membrane. Probes used in Northern blotting can be RNA, DNA, or oligonucleotides of 25 complementary base pairs to the target RNA. They can be complimentary, to the whole RNA or part of the RNA of interest.
Northern blotting helps to analyze a particular gene’s expression levels during morphogenesis and differentiation. The analysis of gene expression is generally, done by microarrays. However, Northern blot analysis can detect small changes in gene expression that microarrays can’t. Northern blot gives information about the size of the mRNA and any precursors.
You may also like:
- Southern blotting: Detection of specific DNA sequence
- Western blotting: Identifying a specific protein

I, Swagatika Sahu (author of this website), have done my master’s in Biotechnology. I have around fourteen years of experience in writing and believe that writing is a great way to share knowledge. I hope the articles on the website will help users in enhancing their intellect in Biotechnology.



